
A Deep And Fuzzy Dive Into Search
Estimated reading time: 16 minutes
Together with my team, I took a deep dive into the available fuzzy search approaches and algorithms for quite a while, in order to find a performant solution for the various projects ArangoSearch gets used for.
Since the introduction of ArangoSearch back in 2018, many of our users have asked for fuzzy search support. We worked hard on getting this done, and the whole team is now excited to finally make fuzzy search available with the upcoming ArangoDB 3.7.
In the following, we will share our learnings and hope they are useful for you.
What is Fuzzy Search?
Nowadays people have to deal with tons of unstructured data. Web-search engines taught us that being inaccurate when searching something is normal, e.g. one can occasionally
- make some typos using a mobile phone and expect typos will be automatically corrected.
- Scientists often deal with the necessity of non-exact matching, e.g. identification of common cognates from different dictionaries is very important in historical linguistics.
- Bioinformatics deals with variations in DNA sequence encoded as an insanely long string of 4 nucleotides (ACTG) and needs some means for quantification.
“Fuzzy search” is an umbrella term referring to a set of algorithms for approximate matching. Usually such algorithms evaluate some similarity measure showing how close a search term is to the items in a dictionary. Then a search engine can make a decision on which results have to be shown first.
In this article I’m going to describe and explain two very important search algorithms, which at the same time are quite different:
- Approximate matching based on Levenshtein distance
- Approximate matching based on NGram similarity
I’m going to dive into details of each algorithm and describe issues one may face when implementing it at web scale. I also emphasize differences between these two approaches so one can better understand various senses of “fuzzy search”.
Approximate Matching Based on Levenshtein Distance
The Levenshtein distance between two words is the minimal number of insertions, deletions or substitutions that are needed to transform one word into the other.
E.g. Levenshtein distance between two words foo and bar is 3, because we have to substitute each letter in word foo to transform it to bar:
- foo -> boo
- boo -> bao
- bao -> bar
Formally, the Levenshtein distance between two strings a and b (of length |a| and |b| respectively) is given by leva,b(|a|, |b|) where leva,b(i, j) is the distance between the first i characters of a and the first j characters of b:
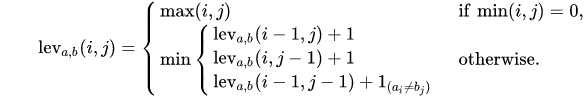
For simplicity let’s assume that the Levenshtein distance is our relevance measure for fuzzy matching, which turns our goal into finding the closest (in terms of Levenshtein distance) items for a given input.
The well known algorithm by Wagner and Fisher uses a dynamic programming scheme that leads to quadratic time complexity in the length of the shortest input string. Of course such complexity makes it impossible to use it at web scale given real world dictionaries may contain hundreds of thousands of items.
In 2002 Klaus Schulz and Stoyan Mihov posted a paper that described a very interesting approach, showing that for any fixed distance n and input sequence W of length N we can compute a deterministic automaton A(W) accepting all sequences with Levenshtein distance less or equal than n to input sequence W in linear time n and space N. The paper itself is rather complex, so I will try to provide an approximation of its main concepts and algorithms. Here I assume you’re familiar with the basics of automata theory.
Once the Levenshtein automaton A(W) is built, we can then intersect it with a dictionary. Of course the complexity of this operation depends on the actual dictionary implementation (e.g. trie, B-tree), but let’s consider a short example. Let’s imagine our dictionary is implemented as trie and stores words “avocado”, “avalon”, “avalanche” and “cargo”:
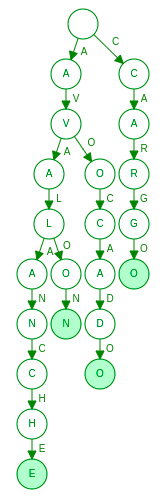
Suppose we search this dictionary for the words within Levenshtein distance 1 of “kargo”. Instead of linearly checking each word in the dictionary, the search algorithm can detect that the whole right branch from root is irrelevant for the given query and completely skip it.
Let’s formalize our task once again:
Given the input sequence W of length N and the maximum acceptable edit distance n we have to build a deterministic finite automaton (DFA) accepting sequence V IFF Lev(W, V) <= n.
In order to better understand how to build a resulting DFA and how it actually works, we’re going to start with a simple example and first draw a nondeterministic finite automaton (NFA) for input sequence foobar and distance 1:
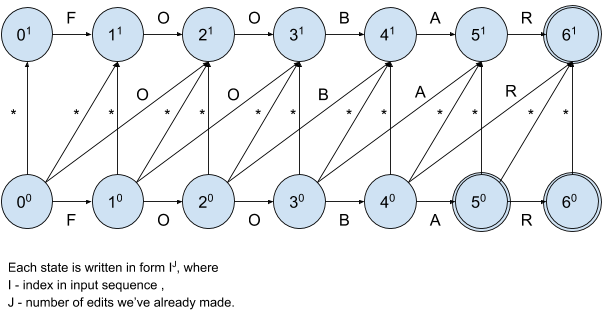
Note that evaluation of NFA is considerably more expensive than the evaluation of DFA since we need to check multiple transitions matching the same label and thus multiple paths from a given state, that’s why we use NFA only to make our explanation clearer.
We start at state 00, horizontal transitions denote correctly entered symbols, verticals and diagonals handle invalid ones. It’s also clear that we’re allowed to pass at most 1 invalid symbol. Whenever the matching encounters an invalid symbol, the NFA flow immediately transfers to the “upper lane” with horizontal transitions only.
Let’s analyze transitions from initial state:
00 -> 01 denotes insertion of any symbol before position 0
00 -> 11 denotes substitution of a symbol at position 0 with any symbol
00 -> 10 denotes correctly entered symbol at position 0
00 -> 21 denotes deletion of a symbol at position 0
Let’s draw the NFA for the same input and a max edit distance of 2:
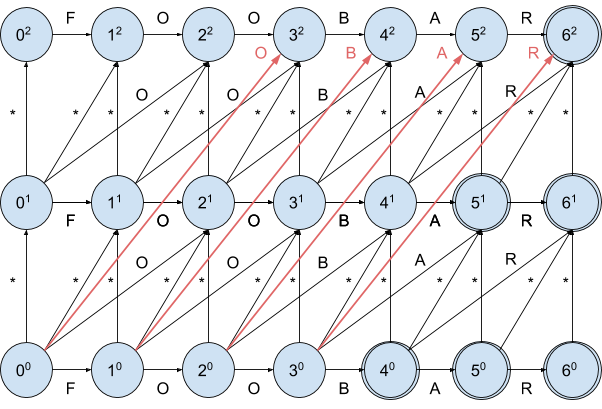
As you can see, the initial state got one more outgoing transition
00 -> 32 denoting pretty much the same as the 00 -> 21 transition, namely the deletion of 2 consequent symbols starting at position 0.
For a max edit distance of 3 there will be 2 more (5 in total) outgoing transitions and so on. That means for each state we have O(n) outgoing transitions.
Blind estimation of number of states in corresponding DFA using the well-known powerset construction will give us O(2(n+1)*N) states which definitely sounds too rough given the shape of NFA.
Schulz and Mihov also showed that given a max edit distance n, position i, and sequence W, we have to analyze at most 2n+1 NFA states. The intuition behind this magic 2n+1 is quite simple: starting from the i-th character of input sequence W, you can’t reach a position higher than (i+n)-th while inserting characters, similarly with the position (i-n)-th while deleting characters.
Let’s improve our estimation based on the observations we’ve just made:
Given the input sequence of length N and max edit distance n, for each character in the input sequence we have to analyze at most 2n+1 NFA states with O(n) outgoing transitions each, i.e. overall number of DFA states is O(n2n+1N) which is definitely better than blind estimation but still too much.
The next important thing which can help us to improve our estimation is the subsumption of relationships between states. As stated in the paper, state IJ subsumes (I+K)J-K, so we don’t need to inspect all possible 2n+1 states. If we now estimate the number of DFA states we get O(n2N).
This estimation is much better than the previous one, but the major issue is its dependency on the input length. As we’ve already noticed, NFA has pretty much the same logical transitions for every state. Let’s generalize it:
| I J -> I J+1 | denotes insertion of a symbol before position I |
| I J -> (I+1) J+1 | denotes substitution of a symbol at position I with any symbol |
| I J -> (I+1) J | denotes correctly entered symbol at position I |
| I J -> (I+K) J+K-1 | denotes deletion of K consecutive symbols after position I |
Such transitions exclusively depend on the distribution of characters with respect to the current character at position I defined as so called characteristic vectors. The characteristic vector X(c, W, I) for character c is a bit set of size min(2n + 1, |W|-I) where the K-th element is equal to 1 IFF Wi+k is equal to c, 0 otherwise.
E.g. for the sequence foobar and distance 1, we have the following characteristic vectors:
X(‘f’, ‘foo’, 0) = { 1, 0, 0 }
X(‘o’, ‘foo’, 0) = { 0, 1, 1 }
X(‘o’, ‘foo’, 2) = { 1, 0, 0 }
As stated above, only 2n+1 states matter for a given position and max edit distance n, thus for each position we can enumerate all 22n+1 transitions over all relevant characteristic vectors (character distributions):
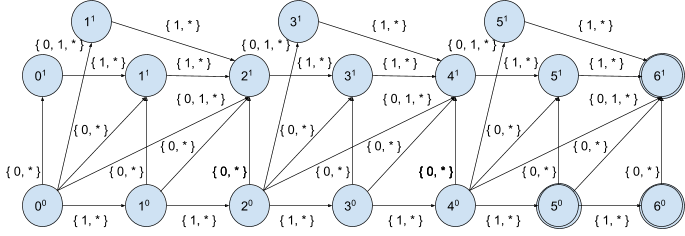
Let’s write it down in tabular form for position 00 and max edit distance 1:
| Characteristic vector X(c, W, i), i < |W| | Set of target positions |
|---|---|
| <0,0,0> | { 01, 11 } |
| <0,0,1> | { 01, 11 } |
| <0,1,0> | { 01, 11, 21 } |
| <0,1,1> | { 01, 11, 21 } |
| <1,0,0> | { 10 } |
| <1,0,1> | { 10 } |
| <1,1,0> | { 10 } |
| <1,1,1> | { 10 } |
And repeat the same exercise for position 01:
| Characteristic vector X(c, W, i), i < |W| | Set of target positions |
|---|---|
| <0,0,0> | { } |
| <0,0,1> | { } |
| <0,1,0> | { } |
| <0,1,1> | { } |
| <1,0,0> | { 11 } |
| <1,0,1> | { 11 } |
| <1,1,0> | { 11 } |
| <1,1,1> | { 11 } |
As you may notice so far we got 5 different sets of target positions:
{ }, { 10 }, { 11 }, { 01, 11 }, { 01, 11, 21 }
By keep enumerating all reachable target states from each position for every relevant character distribution we can see that every position falls down into the one of the following sets denoting states of the corresponding DFA:

Now we can define a generic transition function Δ for max distance 1 describing all possible DFA transitions over character distributions (characteristic vectors). In tabular form it looks like:
| X(c, W, I), 0 <= I <= |W| -3 | AI | BI | CI | DI | EI |
| { 0, 0, 0 } | CI | ∅ | ∅ | ∅ | ∅ |
| { 0, 0, 1 } | CI | ∅ | ∅ | BI+3 | BI+3 |
| { 0, 1, 0 } | EI | ∅ | BI+2 | ∅ | BI+2 |
| { 0, 1, 1 } | EI | ∅ | BI+2 | BI+3 | CI+2 |
| { 1, 0, 0 } | AI+1 | BI+1 | BI+1 | BI+1 | BI+1 |
| { 1, 0, 1 } | AI+1 | BI+1 | BI+1 | DI+1 | DI+1 |
| { 1, 1, 0 } | AI+1 | BI+1 | CI+1 | BI+1 | CI+1 |
| { 1, 1, 1 } | AI+1 | BI+1 | CI+1 | DI+1 | EI+1 |
Schulz and Mihov generalized transition function Δ for arbitrary max distance n so that created it once, we can instantiate any DFA A(W) in linear time O(|W|). Instead of O(n2N) we now have just O(N) linear time complexity, and that’s amazing!
Schulz and Mihov also revealed the very interesting fact that with just a minor addition to the transition function Δ we can easily support Damerau-Levenshtein distance, which treats transpositions atomically, e.g. given two words foobar and foobra we have 2 for Levenshtein distance, but 1 for Damerau-Levenshtein. I’d encourage you to check the original paper if you’re interested in details.
Let’s put it all together
In order to generate the DFA for an input sequence W of length N and max edit distance n in linear time O(N) we have to:
- Define a transition function Δ for a distance n
- For each character in the input sequence use Δ to generate a set of valid transitions
Caveats
As you may already notice, the number of transitions in a transition function grows quadratically O(n2), e.g. we have have
- 5*23=40 transitions for n=1
- 30*25=192 transitions for n=2
- 196*27=25088 transitions for n=3
- 1353*29=692736 transitions for n=4
That quadratic growth makes this approach infeasible for higher distances, thus in ArangoSearch we limit maximum edit distance to 4 and 3 for Levenshtein and Damerau-Levenshtein cases correspondingly.
Schulz and Mihov proved that the edit distance between two sequences W and V is the same as for their corresponding reversed representations W’ and V’, i.e. Lev(W, V) = Lev(W’, V’).The idea is to manage 2 term dictionaries for original and reversed terms (so called FB-Trie). Given that, we can use 2 smaller DFAs for a max distance n in order to look up terms at a max distance n+1 which makes perfect sense given the exponential growth of DFA states with respect to n.
Why another kind of fuzziness?
In real world applications using plain edit distance usually isn’t enough, we need something which can show us how this particular sequence is malformed with respect to the input. An obvious solution is normalization by sequence length, e.g. we can define Levenshtein similarity function as LevSim(W, V) = Lev(W, V) / min(|W|, |V|). But given we are restricted by the max supported edit distance that can be efficiently evaluated, long sequences will always produce LevSim(W,V) values which are close to 0.
Another example is fuzzy phrase search. Of course we can allow to have at most n edits for every word in a phrase, making it possible to have typos in each.
For example, a max distance 1 query with “quck brwn fx” input can match “quick brown fox”, but not “quick-witted brown fox”. Let’s consider another approach of fuzzy search which can help us to serve such queries.
Approximate matching based on n-gram similarity
Another way of calculating similarity between two sequences is calculating the longest common sequence (LCS) of characters. The longer the LCS is, the more similar are the specified sequences. But this approach has one big disadvantage, namely the absence of context. For example, the words connection and fonetica have a long LCS (o-n-e-t-i) but very different meanings.
To overcome this disadvantage, Grzegorz Kondrak described a set of modifications to LCS-based similarity in a paper . The major idea is to use n-gram sequences instead of single characters to carry some more context around. For those who are not familiar with the notion of n-gram, it’s just a contiguous sequence of n items from a given sequence. We can split each sequence into a series of subsequences of length n and use them to determine the LCS.
If we use the same words (connection and fonetica), but calculate similarity based on 3-grams we will get a better similarity measure, e.g. con-onn-nne-nec-ect-cti-tio-ion vs. fon-one-net-eti-tic-ica gives a shorter LCS (zero matches).
Formally it can be defined as follows:
Let X = { x1, x2 … xk } and Y = { y1, y2 … yl } be sequences of length k and l, respectively, composed of symbols of a finite alphabet.
Let Γi,j = (x1 … xi , y1 … yj) be a pair of prefixes of X and Y , and Γ*i,j = (xi+1 … xk , yj+1 … yl ) a pair of suffixes of X and Y.
Let Γni,j = (xi+1 … xi+n , yj+1 … yj+n) be a pair of n-grams in X and Y. If both sequences contain exactly one n-gram, our initial definition is strictly binary: 1 if the n-grams are identical, and 0 otherwise.
Then n-gram similarity sn can be defined as:
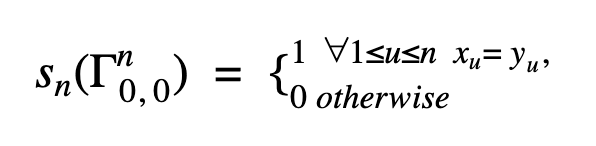
Consequently for sequences X and Y n-gram similarity is recursively defined as:
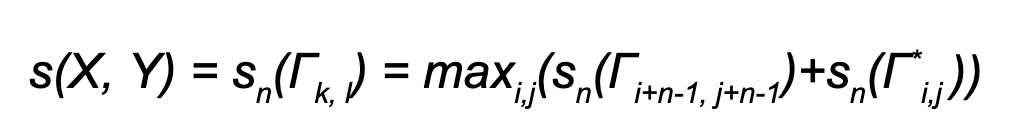
It’s also worth normalizing the LCS by word length to get a value in range [0;1] and avoid the length bias:
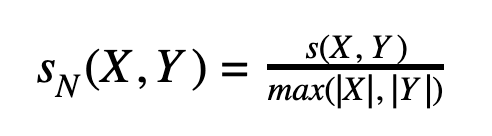
In contrast to Levenshtein-based fuzzy search, this approach can be efficiently implemented in a search engine without building any special structures at query time. We only need to split an input sequence into a set of n-grams, store them as terms in a term dictionary and track positions of each term within a document.
At query time we split a specified input into a set of n-gram tokens and lookup postings for each of them. Then for each matched document we utilize positional information to evaluate the LCS of n-grams and finally calculate sN(X, Y).
Further improvements
To improve search quality, Kondrak suggested an interesting addition to the described approach. He suggested to count identical unigrams in corresponding positions within n-grams:
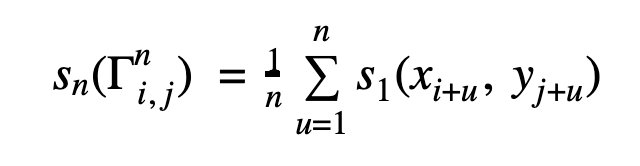
Depending on the use case he got 10-20% more accurate results with the refined approach.
Unfortunately the implementation is not as trivial as the original one, but having a Levenshtein automaton definitely helps: we can split a query into a set of n-gram tokens as in the previous example. For each token W we instantiate a Levenshtein automaton A(W) and use them to get the variations of n-grams from the term dictionary. Given the number such n-gram tokens might be relatively high, it’s definitely worth to union them into the one big automaton which can be used to query the term dictionary efficiently. Then for each matched n-gram we calculate the Levenshtein distance and measure unigram similarity based on edit distance and get more accurate results.
Demo
Though this article was intended to be mostly theoretical, I can imagine many people are eager to try out the described approaches with ArangoSearch. For curious readers we’ve prepared a Jupyter Notebook with an interactive tutorial. The tutorial runs on ArangoDB’s cloud service ArangoGraph works data from the International Movie Database (IMDB)
Tutorial: Fuzzy Search Approaches in ArangoSearch
You can also dive a bit deeper into the Architecture of ArangoSearch, the query capabilities or browse the documentation.
Summary
In this article I did my best to provide a comprehensive and simple explanation of the background behind fuzzy search techniques we implicitly use in our day to day life.
As usual there is no silver bullet and one has to carefully select the most suitable approach for the actual use case or even combine multiple techniques.
I hope you found this interesting and useful.
References
- https://en.wikipedia.org/wiki/Wagner%E2%80%93Fischer_algorithm
- https://en.wikipedia.org/wiki/Trie
- https://en.wikipedia.org/wiki/Nondeterministic_finite_automaton
- https://en.wikipedia.org/wiki/Deterministic_finite_automaton
- https://en.wikipedia.org/wiki/Damerau%E2%80%93Levenshtein_distance
- https://en.wikipedia.org/wiki/N-gram
- https://en.wikipedia.org/wiki/Longest_common_subsequence_problem#Second_property
- http://citeseerx.ist.psu.edu/viewdoc/summary?doi=10.1.1.16.652
- http://www.cis.uni-muenchen.de/people/Schulz/Pub/aspaperCISreport.pdf
- http://citeseerx.ist.psu.edu/viewdoc/download?doi=10.1.1.67.9369&rep=rep1&type=pdf
Continue Reading
ArangoDB 3.5 RC1: Graph Database Improvements with PRUNE & k Shortest Paths
Get the latest tutorials, blog posts and news:
 Skip to content
Skip to content 